27-Aug-2024
The molecular mechanisms governing the regulation of gene expression are of significant interest to biological researchers across a large number of fields. An important cog in the gene regulation machinery is the 1.5MDa multisubunit complex dubbed Integrator. Integrator is responsible for coordinating several activities, such as the 3’ processing of a number of RNAPII derived transcripts including non-polyadenylated small-nuclear RNAs. In addition to this, Integrator is involved in the transcription regulation of a variety of other RNAs, including enhancer RNA, telomerase RNA, long non-coding RNA, and messenger RNA.
As one might expect for a MDa complex, Integrator forms interactions with many other transcriptional regulators, and due to its role in regulating promoter proximal pausing during the transcription of protein coding genes it is regarded as a vital component of the gene-regulation machinery. Despite its important function, Integrator is not completely understood from a structural perspective, with the ‘arm module’ region (INTS 10/13/14/15) remaining poorly understood from a structural perspective. In this study, the INTS10/13/14/15 arm module was expressed and purified from insect cells prior to cryo-EM analysis, yielding a 3.3Å structure of the elongated module with a hook-like structure formed from the INTS13/INTS14 subunits at one end of the structure (Fig 1A).
To understand how the arm module fits within the entire Integrator complex, the authors studied another Integrator subcomplex, the INTS 5/8/10/15. This subcomplex possesses two subunits found within the arm module (INTS10/15), and two found in a previously solved structure of the Integrator complex that lacked the arm module (INTS5/8). Cryo-EM analysis of the INTS5/8/10/15 subcomplex revealed a 3.2Å elongated structure, with INTS15 bridging INTS5/8 and INTS10 (Fig 1A). A key interaction interface between INTS5 and INTS15 was validated by pulldowns from HEK293 cells and Mass Photometry.
By virtue of the overlapping subunits in these two complexes, it was possible to model the arm region in a previously generated structure depicting Integrator bound to RNAPII paused on DNA (Fig 1B/C). Remarkably the modelled position of the arm region aligned with a region of unassigned density in this structure. The arm region is close to the DNA upstream of the pause site, which could facilitate a direct interaction between the two, or bring interacting transcription factors into proximity with the DNA.
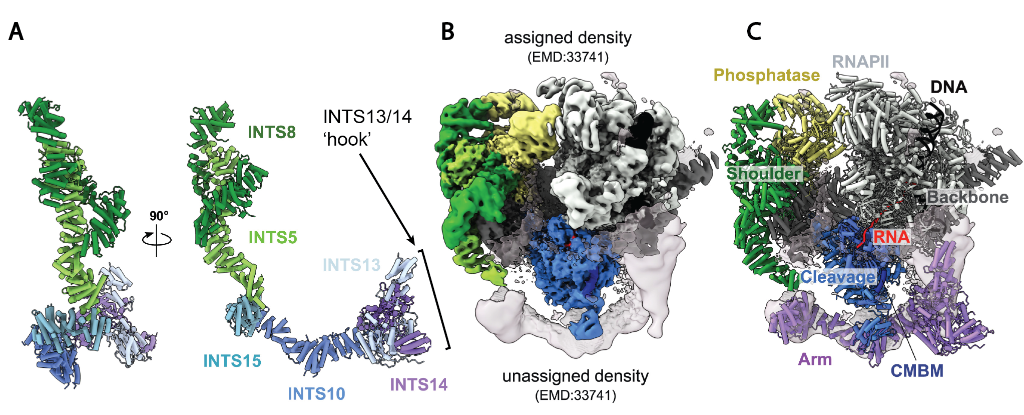
Figure 1: Structural studies of Integrator subcomplexes. 1A: Combined structure of the Integrator Arm module and INTS5/8/10/15. Figure 1B: Previously generated structure depicting integrator subunits bound to RNAPII interacting with DNA, unassigned density is also labelled. 1C: The Arm module fits well into the unassigned density present in 1B.
Several transcription factors are already known to interact with Integrator. To build on our understanding of this, the authors set up a high throughput screen against INTS13 using AlphaFold2 (AF) and a database of over 1500 transcription factors (Figure 2A). The pTM/ipTM (predicted template modelling, and integrated predicted template modelling) and PAE (Predicted alignment error) metrics associated with each predicted structure give an indication on how confident AF is on the predicted structure. By comparing these metrics (Figure 2B) potential interactors were identified, with 25 transcription factors taken forward for further investigation based on their ipTM scores. Analysis of the interactions between these transcription factors revealed two major binding sites: the INTS13 VWA domain, and the INTS13 beta-barrel (Fig 2C).
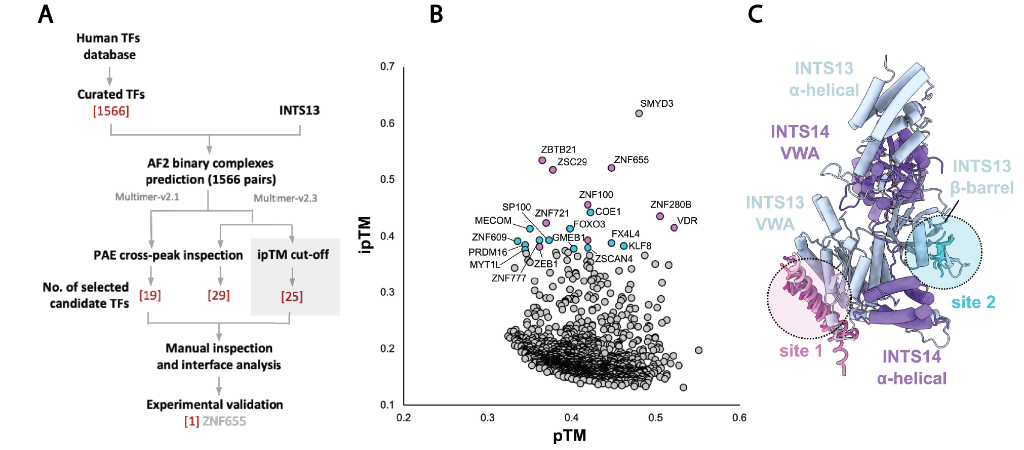
Figure 2: Output of the AF interaction screen. 2A: Workflow overview. 2B: Comparison of pTM and ipTM scores for transcription factors screened, 25 selected transcription factors are highlighted. 2C: Schematic showing transcription factor interaction sites on INTS13.
One of the strongest hits, ZNF655, had previously been shown to copurify with INTS13/14 and as such was chosen for further validation. A reversal of the AF screen found that ZNF655 only interacted with INTS13, and that mutations of key residues at the predicted interface of the two proteins abrogated the interaction in vivo. The flexibly positioned Zn2+ fingers within ZNF655 cover a region of 260A, allowing them to reach regions of DNA up or downstream of the pause site, as well as nascent RNAPII produced RNA. Interestingly, the abrogation of the interaction by mutating key interface residues also led to an almost complete loss of the entire integrator complex, demonstrating its functional significance, and importance to the complex.
Through Instruct-ERIC, you can utilise a fully funded pipeline of structural biology facilities similar to those used by the authors of this paper. Insect-cell expression, biomolecular characterisation including mass photometry, and cryo-EM are all available alongside expert advice at Instruct-ERIC Centres. Discussions regarding the provision of the expertise required for the use of specialised computational structural biology workflows, such as the AlphaFold interaction screen shown here, are currently ongoing!