EOSC Activities and Projects
What is EOSC?
The European Open Science Cloud (EOSC) is an environment, governed by the EOSC Association, meant to federate existing research data infrastructures in Europe and realise a web of FAIR data and related services for science, making research data interoperable and machine actionable following the FAIR guiding principles.
Involvement of Instruct-ERIC in EOSC-related activities and projects
Instruct-ERIC is a partner in the EOSC Association and several EOSC-related projects such as EOSC-Life, providing valuable insight and assistance in developing the EOSC environment. As structural biology is an integral component of the life science network, Instruct’s contribution to the EOSC can provide meaningful benefits to the cloud usage of the European science community.
EOSC-Life
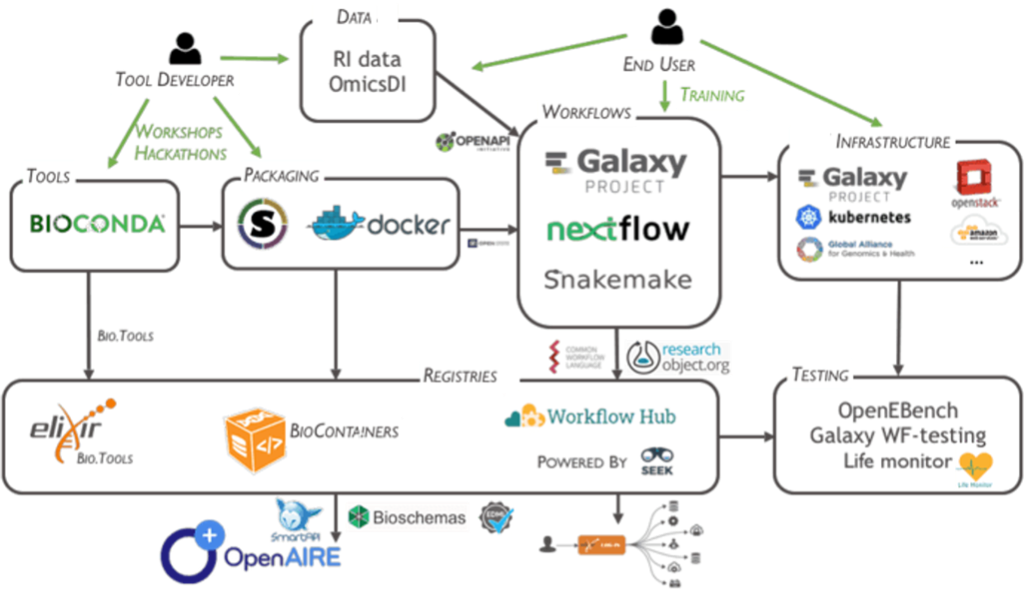
EOSC-Life brings together 13 Life Science Research Infrastructures (RIs) to create an open, digital and collaborative space for data resources and data processing workflows in the European Open Science Cloud (EOSC).
Key resources and services developed by EOSC-Life:






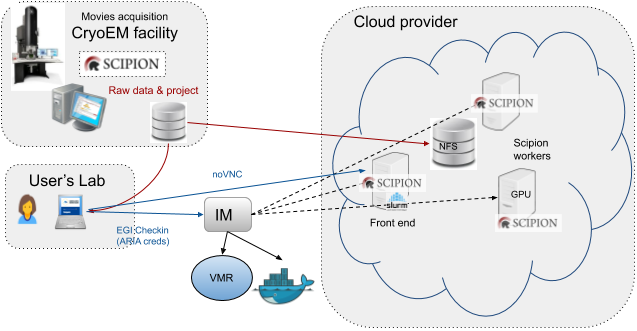 Scipion is a workflow management system developed by I2PC in Madrid that integrates many important software packages available to process CryoEM data.
Scipion is a workflow management system developed by I2PC in Madrid that integrates many important software packages available to process CryoEM data. 






