21-Oct-2025
Single-stranded DNA-binding proteins (SSBs) coat single-stranded DNA (ssDNA) and are fundamental to DNA replication. In eukaryotes, the main component of SSB is the RPA (Replication Protein A) complex, a key factor in DNA replication by promoting the assembly, binding, and activity of DNA polymerases. RPA also has a variety of other functions in eukaryotes, including DNA repair.
This study from Martínez-Carranza et al (2024), aims to study the function of RPA in archaebacteria, understanding how it interacts with DNA polymerases for replication processes. DNA replication in archaeal chromosomes is similar to eukaryotic examples, including in RPA structure formation.
Previous studies had indicated that the C-terminal region of RPA had a “winged-helix” (WH) domain, shown to be involved in the binding of DNA repair enzymes, and for DNA replication. Using an integrative structural biology approach, the team from Institut Pasteur in Paris, solved the structure of this WH region, to understand how it interacts with archaeal DNA replisomes (PriSL and PolD), conserved in eukaryote organisms.
The use of NMR uncovered the solution structure of the C-terminal region of Rpa2, demonstrating that its structure is a three-helix bundle and a short three-stranded antiparallel β sheet, forming a rigid globular domain. The importance of the WH region of Rpa2 was made clear by deletion attempts, combined with previous studies indicating that Rpa2 was responsible for DNA polymerase binding, rather than direct binding to DNA strands.
Bio-layer interferometry (BLI) indicated that deletion of Rpa2WH disrupted the interaction of the RPA protein with both PriSL and PolD, and NMR found that the globular domain of Rpa2 is responsible for this binding.
X-ray crystallography was used to determine how RPA binds to PriSL. The crystallographic structure of PriSL and the NMR structure of Rpa2WH were reconstructed in an electron density map (Figure 1). Rpa2WH binds to the catalytic subunit of PriSL, and is connected to the core of RPA via a flexible linker. This linker is crucial to the recruitment of PriSL by Rpa2WH, as the RPA trimerisation core is also required, as determined by primer extension assays.
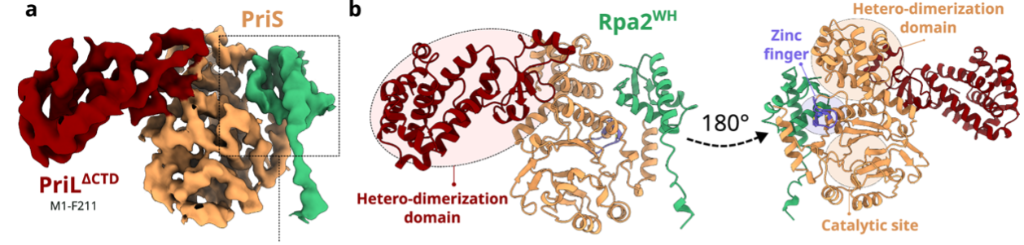
Figure 1. Electron density map and model of the Primase-Rpa2WH complex crystal structure.
To determine the structure of PolD and RPA, cryo-EM was utilised. This was carried out at the Oxford Particle Imaging Centre (OPIC), part of Instruct-UK. PolD is made up of two subunits; one for proofreading (DP1), and one for polymerisation (DP2). The Rpa2WH domain binds with the polymerisation subunit, as shown in Figure 2. Unlike the binding with PriSL, the active site of PolD is far from the binding site with Rpa2WH. Furthermore, the binding surface of both PolD and PriSL with Rpa2WH are very different, with only one residue in common.
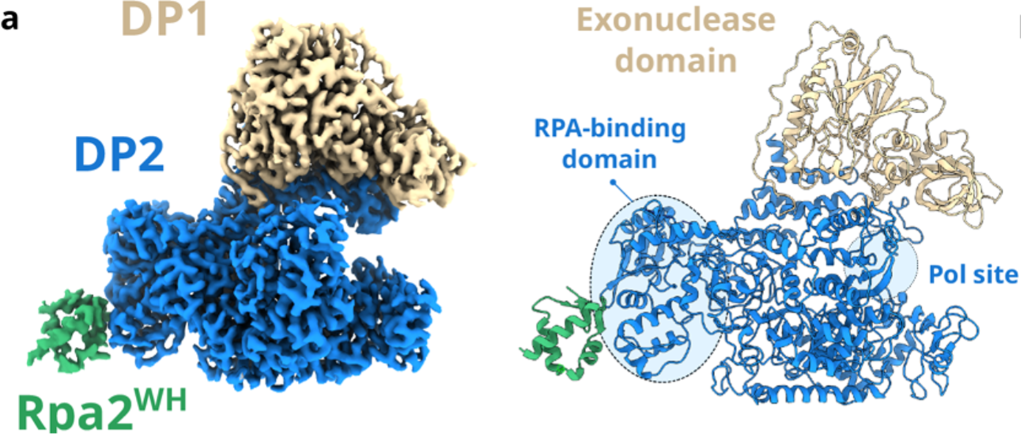
Figure 2. Final composite map of PolD-Rpa2WH at 2.9 Å global average resolution.
A key feature of this study is that there are several conserved binding functions between RPA in archaea, and in humans. The project illustrates the structure of the RPA winged helix binding site with archaea DNA polymerases, but also demonstrates how research into these proteins can be applied to humans downstream, particularly with regard to eukaryotic telomere maintenance and DNA repair.